Unusual Observations
Dr. D’Agostino McGowan
Leverage
- leverage is the amount of influence an observation has on the estimation of ^β
Leverage
- leverage is the amount of influence an observation has on the estimation of ^β
- Mathematically, we can define this as the diagonal elements of the hat matrix.
Leverage
- leverage is the amount of influence an observation has on the estimation of ^β
- Mathematically, we can define this as the diagonal elements of the hat matrix.
What is the hat matrix?
Leverage
- leverage is the amount of influence an observation has on the estimation of ^β
- Mathematically, we can define this as the diagonal elements of the hat matrix.
What is the hat matrix?
- X(XTX)−1XT
Leverage
- leverage is the amount of influence an observation has on the estimation of ^β
- Mathematically, we can define this as the diagonal elements of the hat matrix.
hi=Hiihi=X(XTX)−1XTii
Leverage
What do we use the diagnonal of the hat matrix?
Leverage
What do we use the diagnonal of the hat matrix?
- Recall that the variance of the residuals is
var(ei)=σ2(1−hi)
- so large leverage points will pull the fit towards yi
Leverage
- The leverage, hi, will always be between 0 and 1
How do we know this? Let's show it using the fact that the hat matrix is idempotent and symmetric.
Leverage
- The leverage, hi, will always be between 0 and 1
How do we know this? Let's show it using the fact that the hat matrix is idempotent and symmetric.
hi=∑jHijHji
Leverage
- The leverage, hi, will always be between 0 and 1
How do we know this? Let's show it using the fact that the hat matrix is idempotent and symmetric.
hi=∑jHijHji=∑jH2ij
Leverage
- The leverage, hi, will always be between 0 and 1
How do we know this? Let's show it using the fact that the hat matrix is idempotent and symmetric.
hi=∑jHijHji=∑jH2ij=H2ii+∑j≠iH2ij
Leverage
- The leverage, hi, will always be between 0 and 1
How do we know this? Let's show it using the fact that the hat matrix is idempotent and symmetric.
hi=∑jHijHji=∑jH2ij=H2ii+∑j≠iH2ij=h2i+∑j≠iH2ij
Leverage
- The leverage, hi, will always be between 0 and 1
hi=∑jHijHji=∑jH2ij=H2ii+∑j≠iH2ij=h2i+∑j≠iH2ij
- This means that hi must be larger than h2i, implying that hi will always be between 0 and 1 ✅
Leverage
- The ∑ihi=p+1 (remember when we calculated the trace of H?)
Leverage
- The ∑ihi=p+1 (remember when we calculated the trace of H?)
- This means an average value for hi is (p+1)/n
Leverage
- The ∑ihi=p+1 (remember when we calculated the trace of H?)
- This means an average value for hi is (p+1)/n
- 👍 A rule of thumb leverages greater than 2(p+1)/n should get an extra look
Standardized residuals
- We can use the leverages to standardize the residuals
Standardized residuals
- We can use the leverages to standardize the residuals
- Instead of plotting the residuals, e, we can plot the standardized residuals
ri=e^σ√1−hi
Standardized residuals
- We can use the leverages to standardize the residuals
- Instead of plotting the residuals, e, we can plot the standardized residuals
ri=e^σ√1−hi
- 👍 A rule of thumb for standardized residuals: those greater than 4 would be very unusual and should get an extra look
Application Exercise
| y | x |
|---|---|
| 1 | 0 |
| 5 | 4 |
| 2 | 2 |
| 2 | 1 |
| 11 | 10 |
Using the data above calculate:
- The leverage for each observation. Are any "unusual"?
- The standardized residuals, ri.
Doing it in R
- It is good to understand how to calculate these standardized residuals by hand, but there is an R function that does this for you (
rstandard()) - There is also an R function to calculate the leverage (
hatvalues())
Standardized residuals
mod <- lm(mpg ~ disp, data = mtcars)d <- data.frame( standardized_resid = rstandard(mod), fit = fitted(mod))ggplot(d, aes(fit, standardized_resid)) + geom_point() +geom_hline(yintercept = 0) + labs(y = "Standardized Residual")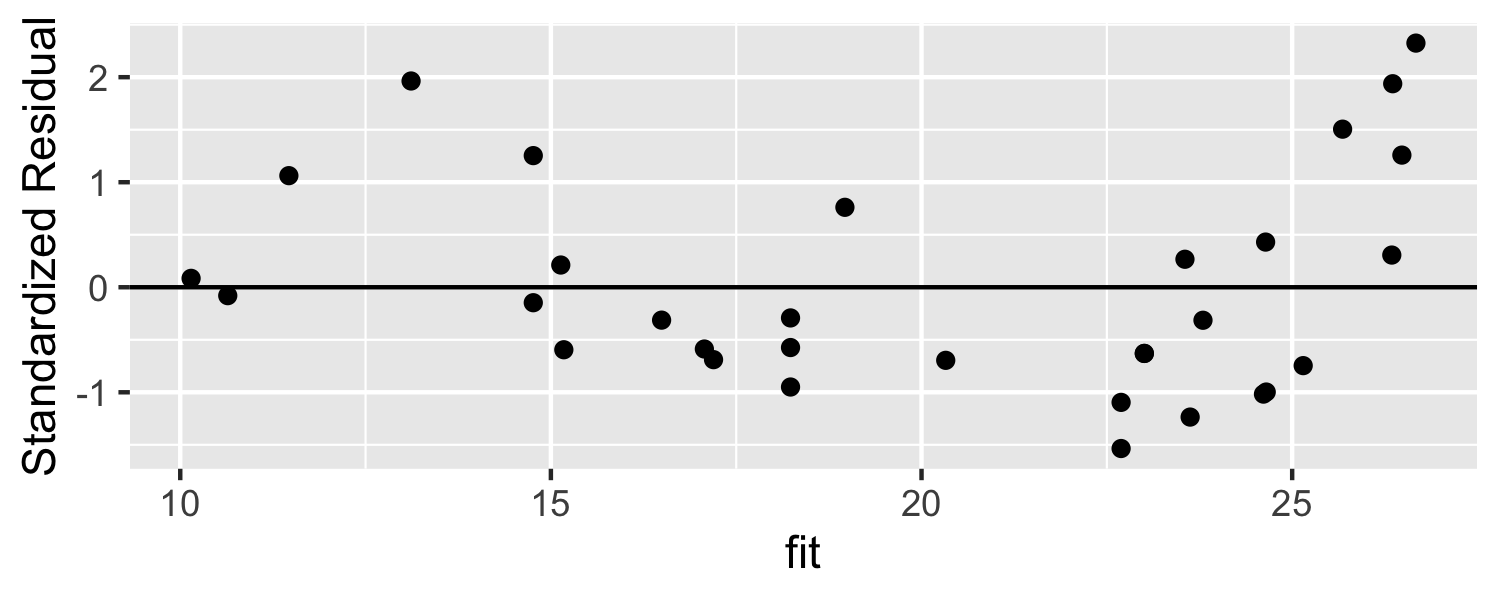
Outliers
- An outlier is a point that doesn't fit the current model well
Outliers
- An outlier is a point that doesn't fit the current model well
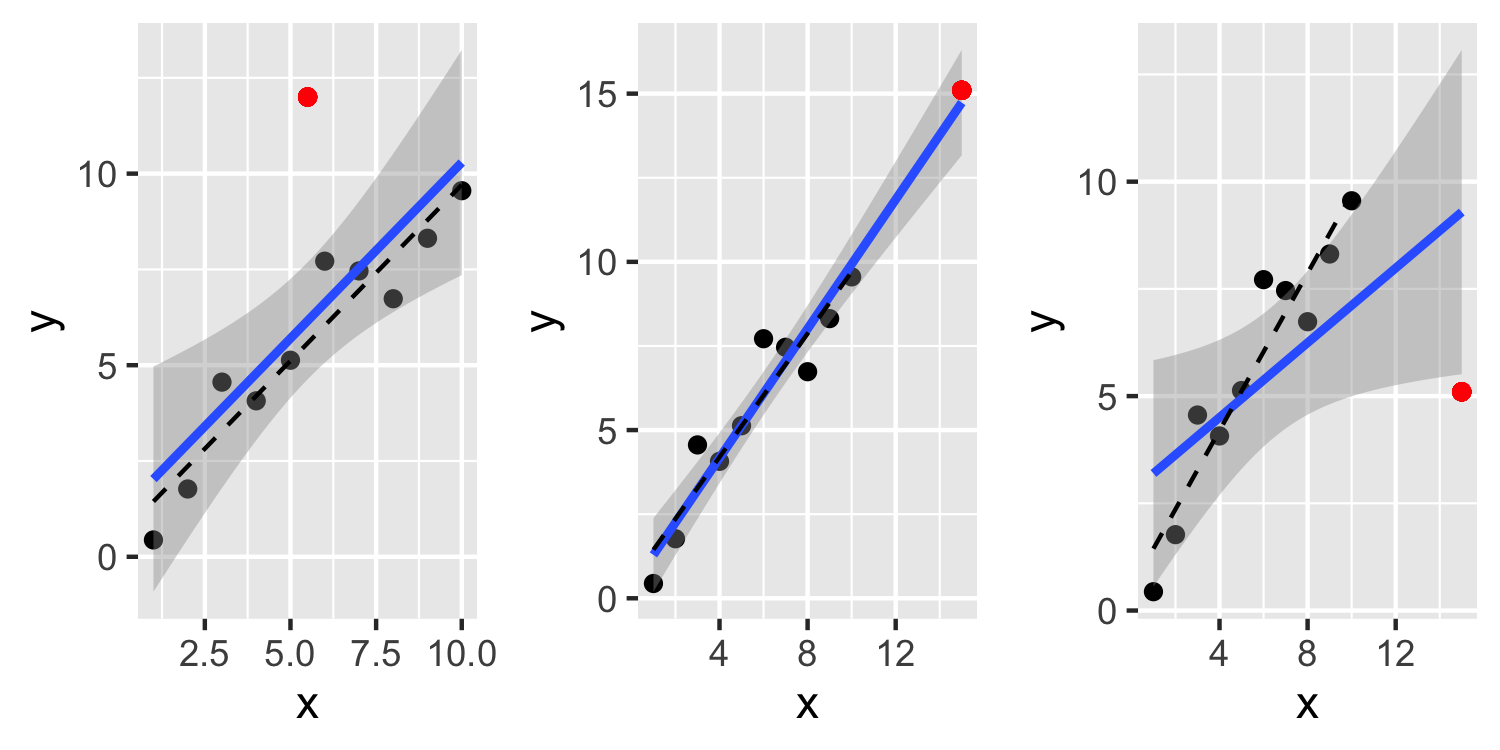
Outliers
- An outlier is a point that doesn't fit the current model well
- This first plot, see a point that is definitely an outlier but it doesn't have much leverage or influence over the fit
Outliers
- An outlier is a point that doesn't fit the current model well
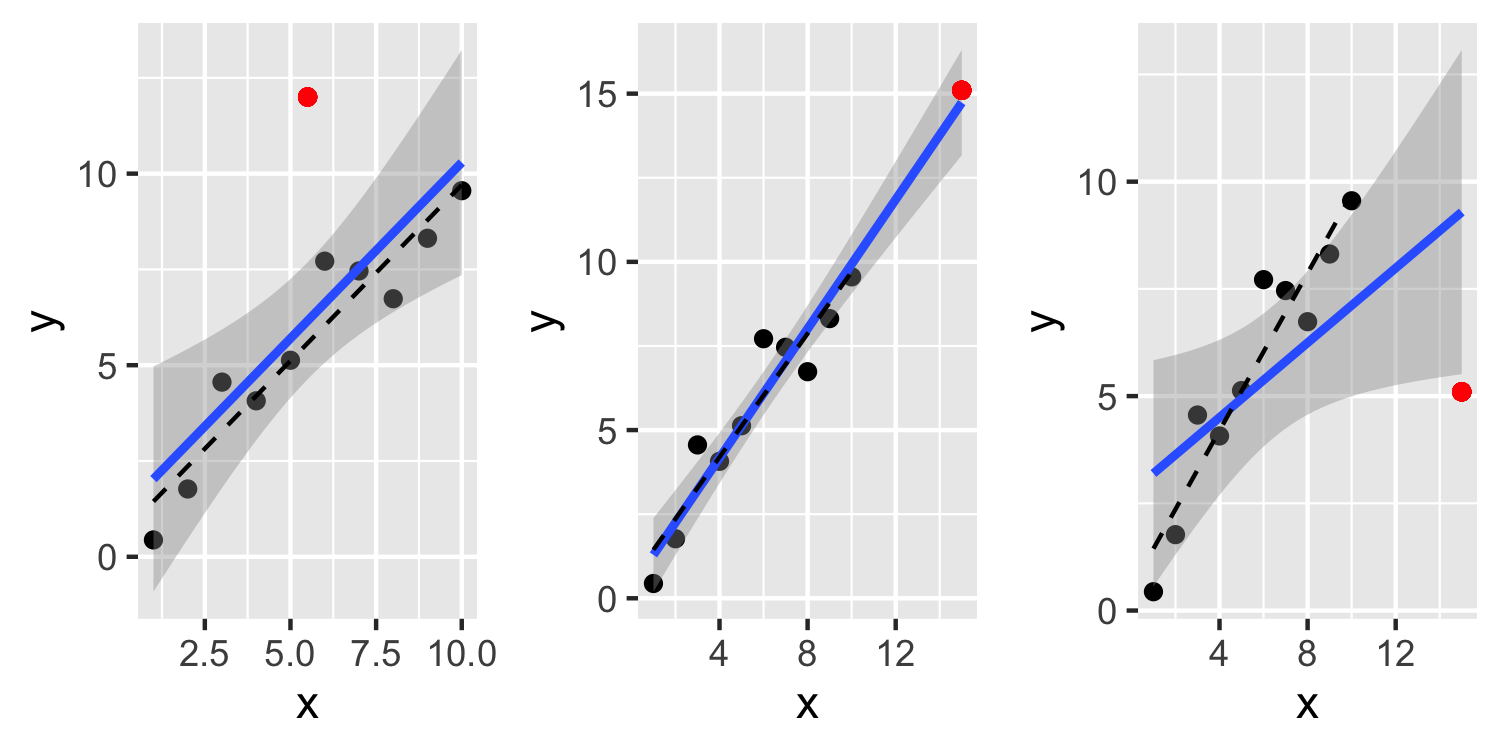
- This second plot, see a point that has a large leverage but is not an outlier and doesn't have much influence over the fit
Outliers
- An outlier is a point that doesn't fit the current model well
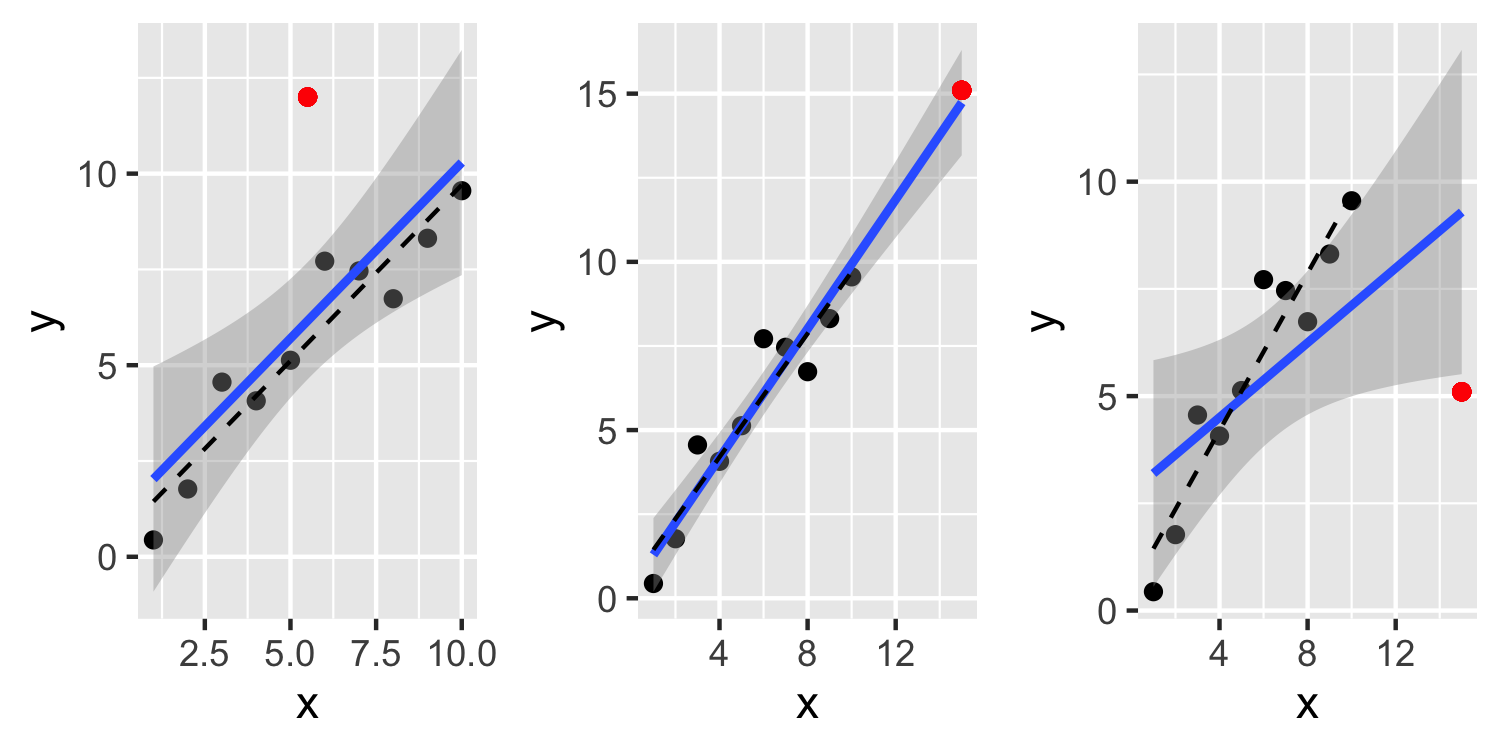
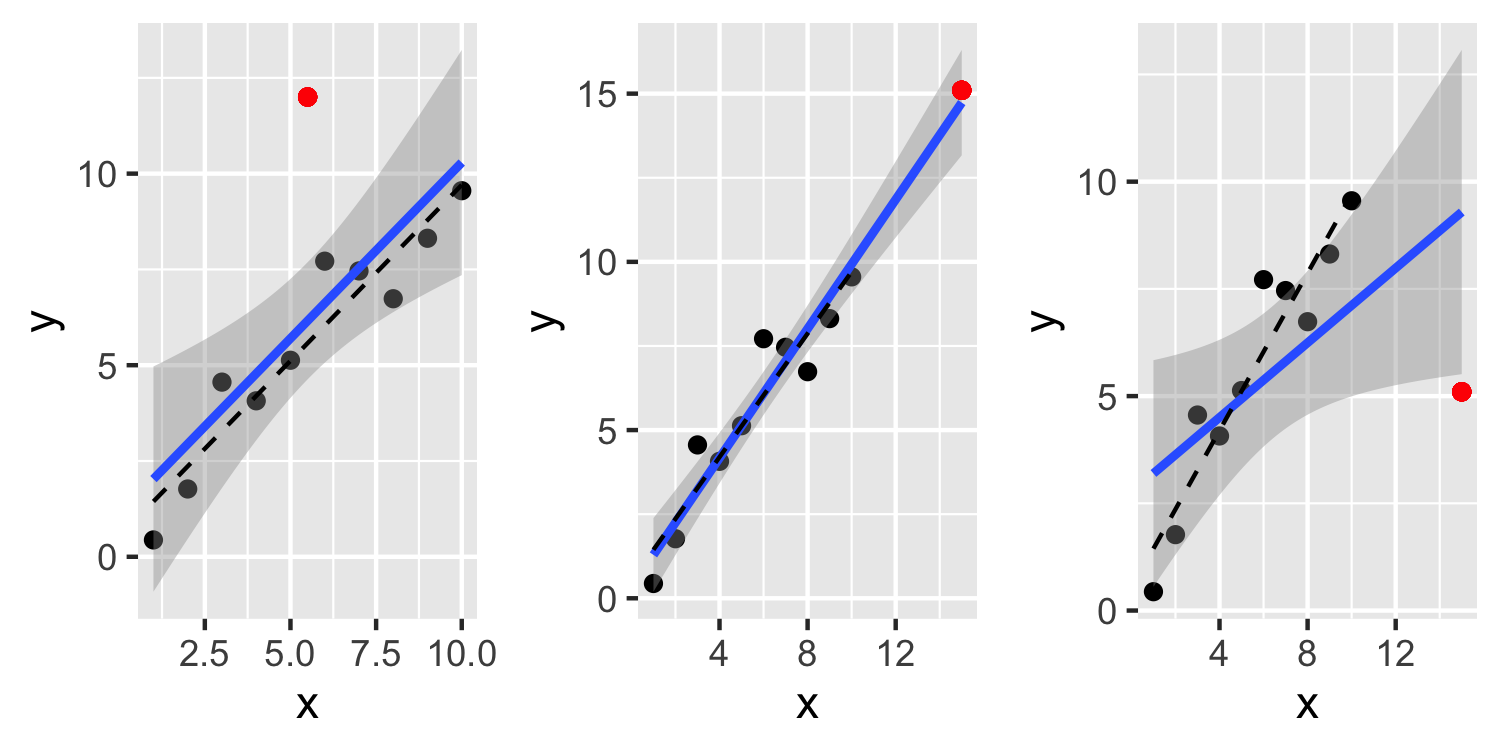
- In the third plot, the point is both an outlier and very influential. Not only is the residual for this point large, but it inflates the residuals for the other points
Outliers
- To detect points like this third example, it can be prudent to exclude the point and recompute the estimates to get ^β(i) and ^σ2(i)
Outliers
- To detect points like this third example, it can be prudent to exclude the point and recompute the estimates to get ^β(i) and ^σ2(i)
^y(i)=xTi^β(i)
Outliers
- To detect points like this third example, it can be prudent to exclude the point and recompute the estimates to get ^β(i) and ^σ2(i)
^y(i)=xTi^β(i)
- If ^y(i)−yi is large, then observation i is an outlier.
Outliers
- To detect points like this third example, it can be prudent to exclude the point and recompute the estimates to get ^β(i) and ^σ2(i)
^y(i)=xTi^β(i)
- If ^y(i)−yi is large, then observation i is an outlier.
How do we determine "large"? We need to scale it using the variance!
Application Exercise
Show that
^var((^y−^y(i)))=^σ2(i)(1+xTi(XT(i)X(i))−1xi)
| y | x |
|---|---|
| 1 | 0 |
| 5 | 4 |
| 2 | 2 |
| 2 | 1 |
| 11 | 10 |
Using the data above, calculate ^var(^y)(i) for observation 5.
Studentized residuals
ti=yi−^y(i)^σ(i)(1+xTi(XT(i)X(i))−1xi)1/2 The have a t distribution with (n−1)−(p+1)=n−p−2 degrees of freedom if the model is correct and ϵ N(0,σ2I).
Studentized residuals
ti=yi−^y(i)^σ(i)(1+xTi(XT(i)X(i))−1xi)1/2 The have a t distribution with (n−1)−(p+1)=n−p−2 degrees of freedom if the model is correct and ϵ N(0,σ2I).
- There is an easier way to compute these using the studentized residuals!
ti=ri(n−p−2n−p−1−r2i)1/2
Application Exercise
| y | x |
|---|---|
| 1 | 0 |
| 5 | 4 |
| 2 | 2 |
| 2 | 1 |
| 11 | 10 |
Calculate the studentized residuals for the data above.
Studentized residuals
It is good to understand how to calculate these studentized residuals by hand, but there is an R function that does this for you (rstudent())
Influential points
- A common measure to determine influential points is Cook's D
Di=(^y−^y(i))T(^y−^y(i))(p+1)^σ2
Influential points
- A common measure to determine influential points is Cook's D
Di=(^y−^y(i))T(^y−^y(i))(p+1)^σ2
- (^y−^y(i)) is the change in the fit after leaving observation i out.
Influential points
- A common measure to determine influential points is Cook's D
Di=(^y−^y(i))T(^y−^y(i))(p+1)^σ2
- (^y−^y(i)) is the change in the fit after leaving observation i out.
- This can be calculated using:
1p+1r2ihi1−hi
Influential points
- A common measure to determine influential points is Cook's D
Di=(^y−^y(i))T(^y−^y(i))(p+1)^σ2
- (^y−^y(i)) is the change in the fit after leaving observation i out.
- This can be calculated using:
1p+1r2ihi1−hi
- 👍 A rule of thumb is to give observations with Cook's Distance > 4/n an extra look
Cook's Distance
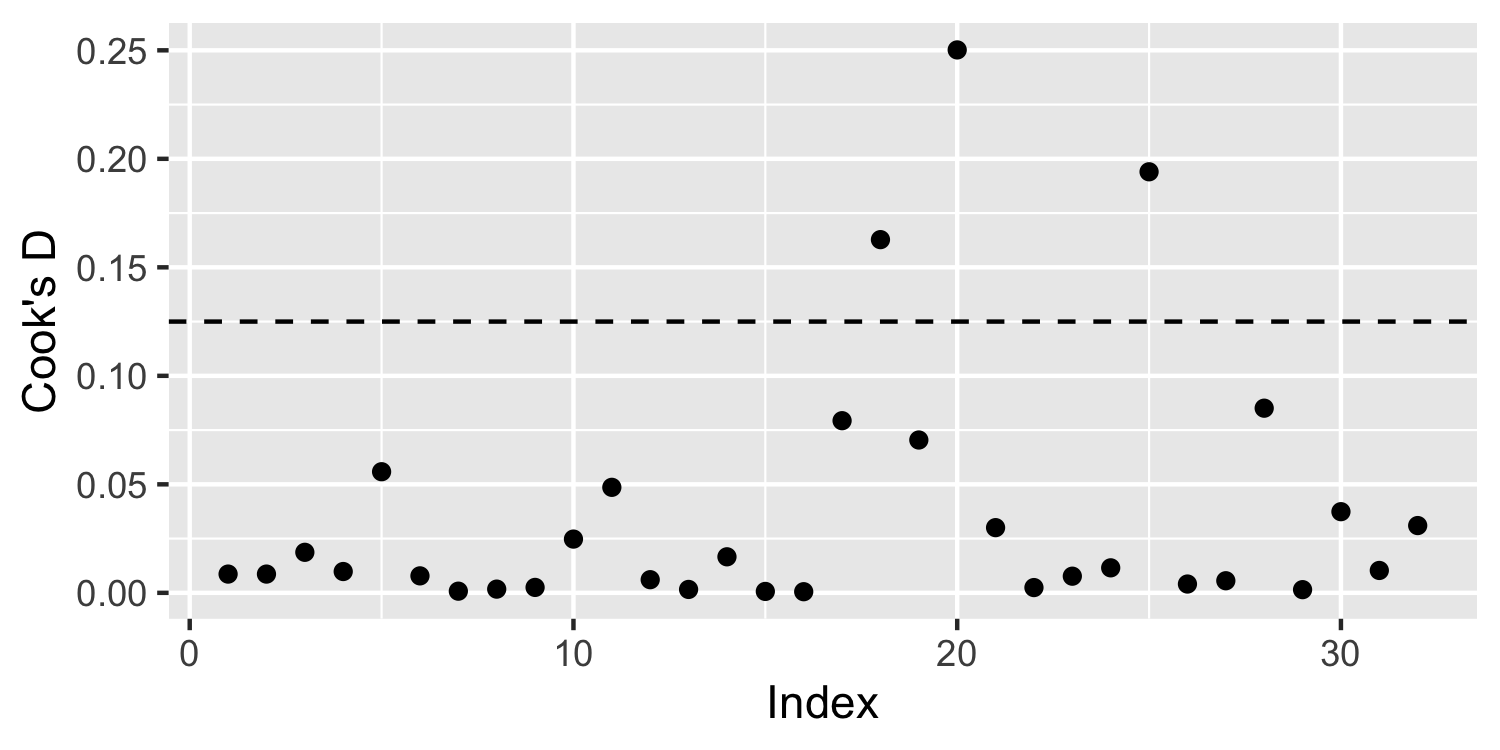
Application Exercise
| y | x |
|---|---|
| 1 | 0 |
| 5 | 4 |
| 2 | 2 |
| 2 | 1 |
| 11 | 10 |
Calculate Cook's Distance for the data above and plot it with the row number on the x-axis and Cook's Distance on the y-axis.
Cook's Distance
It is good to understand how to calculate these by hand, but there is an R function that does this for you (cooks.distance())